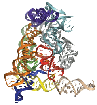Introduction to TargeTron
Mobile group II introns are ribozymes that insert themselves into specific target sites in double-stranded DNA by a distinct mobility mechanism, called retrohoming. The well-studied Lactococcus lactis Ll.LtrB group II intron (Figure 1) consists of an intron RNA and a multifunctional intron-encoded protein (IEP) that has reverse transcriptase (RT), RNA splicing (maturase), and DNA endonuclease (En) activities.
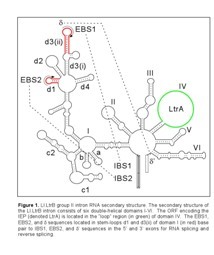 After translation, the IEP binds specifically to the intron in unspliced precursor RNA and promotes formation of the catalytically active RNA structure, which is essential for both intron splicing and intron reverse splicing into a DNA target site. Upon intron splicing, the IEP remains associated with the excised intron lariat RNA to form a ribonucleoprotein complex (RNP) that mediates the mobility of the Ll.LtrB intron (Figure 2). First, the intron RNA reverse splices directly into one strand of the target DNA. The associated IEP then cleaves the opposite strand 9 or 10 nucleotides downstream of the intron insertion site, and uses the cleaved 3' end as a primer for target primed reverse transcription (TPRT) of the inserted intron RNA. The resulting intron cDNA is integrated into genomic DNA by host cell DNA repair mechanisms.
After translation, the IEP binds specifically to the intron in unspliced precursor RNA and promotes formation of the catalytically active RNA structure, which is essential for both intron splicing and intron reverse splicing into a DNA target site. Upon intron splicing, the IEP remains associated with the excised intron lariat RNA to form a ribonucleoprotein complex (RNP) that mediates the mobility of the Ll.LtrB intron (Figure 2). First, the intron RNA reverse splices directly into one strand of the target DNA. The associated IEP then cleaves the opposite strand 9 or 10 nucleotides downstream of the intron insertion site, and uses the cleaved 3' end as a primer for target primed reverse transcription (TPRT) of the inserted intron RNA. The resulting intron cDNA is integrated into genomic DNA by host cell DNA repair mechanisms.
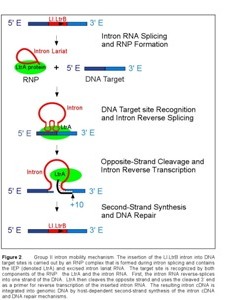 The target site for the Ll.LtrB group II intron extends from position -26 in the 5' exon to position +10 in the 3' exon, as numbered from the intron insertion site (Figure 3). Both components of the RNP complex, the intron RNA and the IEP, are involved in DNA target site recognition. The central sequences of the target site (-12 to -8 and -6 to +2) are recognized primarily by base paring with the intron RNA. The intron RNA sequences (denoted EBS2, EBS1, and δ) involved in this base pairing are located in two different RNA stem loops (Id1 and Id3). The complementary DNA target sequences are denoted IBS2, IBS1, and δ'. The EBS and IBS refer to exon and intron binding sites, respectively. These short sequences are also involved in base-pairing interactions required for intron splicing. The IEP recognizes sequences flanking the central region of the DNA target site. The IEP first recognizes the distal 5' exon region and triggers local DNA unwinding, enabling the formation of base-pairs between the intron and the target site for reverse splicing. Cleavage of the opposite strand occurs after reverse splicing and involves interactions between the IEP and the 3' exon of the target sequence, the most critical being the recognition of T+5.
The target site for the Ll.LtrB group II intron extends from position -26 in the 5' exon to position +10 in the 3' exon, as numbered from the intron insertion site (Figure 3). Both components of the RNP complex, the intron RNA and the IEP, are involved in DNA target site recognition. The central sequences of the target site (-12 to -8 and -6 to +2) are recognized primarily by base paring with the intron RNA. The intron RNA sequences (denoted EBS2, EBS1, and δ) involved in this base pairing are located in two different RNA stem loops (Id1 and Id3). The complementary DNA target sequences are denoted IBS2, IBS1, and δ'. The EBS and IBS refer to exon and intron binding sites, respectively. These short sequences are also involved in base-pairing interactions required for intron splicing. The IEP recognizes sequences flanking the central region of the DNA target site. The IEP first recognizes the distal 5' exon region and triggers local DNA unwinding, enabling the formation of base-pairs between the intron and the target site for reverse splicing. Cleavage of the opposite strand occurs after reverse splicing and involves interactions between the IEP and the 3' exon of the target sequence, the most critical being the recognition of T+5.
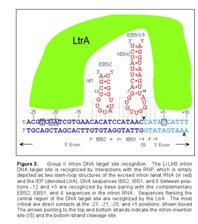 Because two crucial segments of the DNA target site are recognized by base paring interactions with the intron RNA, it is possible to change the specificity of the Ll.LtrB intron by modifying the target site recognition sequences in the intron RNA. This flexibility allowed Ll.LtrB to be developed into a highly efficient bacterial gene targeting vector called a TargeTron. Currently, the Targetrons are designed to insert into specific chromosomal loci by a computer algorithm. First, the algorithm uses the probabilistic model to evaluate a DNA sequence for potential intron insertion sites recognized by the IEP and ranks the target sites using a log-odds score. In that process, each target site residue is assigned a value based on the position-specific nucleotide frequency of that residue in a database of active Ll.LtrB group II intron target sites. Next, PCR primers are designed to modify the introns EBS2, EBS1 and δ sequences for insertion into those sites. If the intron is expressed from a donor plasmid, the IBS sequences in the plasmids 5' exon must also complement the introns target site recognition sequences. This is necessary to ensure efficient intron forward splicing and subsequent RNP formation. The required modifications are introduced by a one-step PCR, using three designed primers and one universal primer to mutate the intron at several positions spanning a 350 bp region.
Because two crucial segments of the DNA target site are recognized by base paring interactions with the intron RNA, it is possible to change the specificity of the Ll.LtrB intron by modifying the target site recognition sequences in the intron RNA. This flexibility allowed Ll.LtrB to be developed into a highly efficient bacterial gene targeting vector called a TargeTron. Currently, the Targetrons are designed to insert into specific chromosomal loci by a computer algorithm. First, the algorithm uses the probabilistic model to evaluate a DNA sequence for potential intron insertion sites recognized by the IEP and ranks the target sites using a log-odds score. In that process, each target site residue is assigned a value based on the position-specific nucleotide frequency of that residue in a database of active Ll.LtrB group II intron target sites. Next, PCR primers are designed to modify the introns EBS2, EBS1 and δ sequences for insertion into those sites. If the intron is expressed from a donor plasmid, the IBS sequences in the plasmids 5' exon must also complement the introns target site recognition sequences. This is necessary to ensure efficient intron forward splicing and subsequent RNP formation. The required modifications are introduced by a one-step PCR, using three designed primers and one universal primer to mutate the intron at several positions spanning a 350 bp region.
In addition to site-specific gene inactivation, Targetrons can be used to deliver genes cloned within the introns domain IV to desired chromosomal locations, to repair mutant genes or to generate double stranded breaks and stimulate homologous recombination. In principle, group II intron RNPs can be delivered into mammalian cells to perform targeted genetic repair, although significant technical issues must still be overcome for the development of any new therapeutic approaches.
In bacteria, the Targetron is commonly expressed from a donor plasmid in which the IEP open reading frame (ORF) has been deleted from intron domain IV. The IEP is expressed separately from a position downstream of the 3' exon and intron RNPs are assembled in the host cell. This arrangement enables achieving very high intron mobility frequencies probably due to greater resistance of the ΔORF intron to nucleolytic cleavage in domain IV. The insertion of the ΔORF intron leads to a gene disruption because the intron cannot splice in the absence of the IEP. In principle, the Targetron can be used to obtain either permanent or conditional gene disruption depending on the orientation of the inserted intron with respect to target gene transcription. A Targetron inserted in the sense orientation is transcribed into a functional intron RNA and can be spliced from a precursor RNA to restore the mRNA by providing the IEP in trans, leading to conditional disruption. A Targetron inserted in the antisense orientation is transcribed into non-functional complementary intron sequence and cannot be spliced, yielding a permanent gene disruption.
The major advantage of using Targetrons for gene targeting in bacteria is that computer-designed Targetrons are extremely efficient that allow mutants to be generated without the use of antibiotic resistance markers. This is crucial when the host is naturally resistant to common antibiotics, or for cases in which government regulations restrict the insertion of new antibiotic resistance genes into bioterrorism-related organisms. Also, when multiple knockouts are desired the selection process can be expedited since Targetrons can be introduced sequentially without requiring the intermediate removal of antibiotic resistance markers.
A new Targetron based on the EcI5 group II intron (which was discovered in Escherichia coli virulence plasmid pO157) has recently been developed. The EcI5 recognizes an entirely different target sequence than the LtrB intron, but can again be programmed to insert specifically into desired chromosomal loci. Computer-designed EcI5 Targetrons have been shown to integrate specifically at ten different sites in the E. coli chromosomal lacZ gene at frequencies up to 98% without selection. Based on these and other experiments, the EcI5 Targetron seems to be more efficient that the LtrB Targetron.
 After translation, the IEP binds specifically to the intron in unspliced precursor RNA and promotes formation of the catalytically active RNA structure, which is essential for both intron splicing and intron reverse splicing into a DNA target site. Upon intron splicing, the IEP remains associated with the excised intron lariat RNA to form a ribonucleoprotein complex (RNP) that mediates the mobility of the Ll.LtrB intron (Figure 2). First, the intron RNA reverse splices directly into one strand of the target DNA. The associated IEP then cleaves the opposite strand 9 or 10 nucleotides downstream of the intron insertion site, and uses the cleaved 3' end as a primer for target primed reverse transcription (TPRT) of the inserted intron RNA. The resulting intron cDNA is integrated into genomic DNA by host cell DNA repair mechanisms.
After translation, the IEP binds specifically to the intron in unspliced precursor RNA and promotes formation of the catalytically active RNA structure, which is essential for both intron splicing and intron reverse splicing into a DNA target site. Upon intron splicing, the IEP remains associated with the excised intron lariat RNA to form a ribonucleoprotein complex (RNP) that mediates the mobility of the Ll.LtrB intron (Figure 2). First, the intron RNA reverse splices directly into one strand of the target DNA. The associated IEP then cleaves the opposite strand 9 or 10 nucleotides downstream of the intron insertion site, and uses the cleaved 3' end as a primer for target primed reverse transcription (TPRT) of the inserted intron RNA. The resulting intron cDNA is integrated into genomic DNA by host cell DNA repair mechanisms.
 The target site for the Ll.LtrB group II intron extends from position -26 in the 5' exon to position +10 in the 3' exon, as numbered from the intron insertion site (Figure 3). Both components of the RNP complex, the intron RNA and the IEP, are involved in DNA target site recognition. The central sequences of the target site (-12 to -8 and -6 to +2) are recognized primarily by base paring with the intron RNA. The intron RNA sequences (denoted EBS2, EBS1, and δ) involved in this base pairing are located in two different RNA stem loops (Id1 and Id3). The complementary DNA target sequences are denoted IBS2, IBS1, and δ'. The EBS and IBS refer to exon and intron binding sites, respectively. These short sequences are also involved in base-pairing interactions required for intron splicing. The IEP recognizes sequences flanking the central region of the DNA target site. The IEP first recognizes the distal 5' exon region and triggers local DNA unwinding, enabling the formation of base-pairs between the intron and the target site for reverse splicing. Cleavage of the opposite strand occurs after reverse splicing and involves interactions between the IEP and the 3' exon of the target sequence, the most critical being the recognition of T+5.
The target site for the Ll.LtrB group II intron extends from position -26 in the 5' exon to position +10 in the 3' exon, as numbered from the intron insertion site (Figure 3). Both components of the RNP complex, the intron RNA and the IEP, are involved in DNA target site recognition. The central sequences of the target site (-12 to -8 and -6 to +2) are recognized primarily by base paring with the intron RNA. The intron RNA sequences (denoted EBS2, EBS1, and δ) involved in this base pairing are located in two different RNA stem loops (Id1 and Id3). The complementary DNA target sequences are denoted IBS2, IBS1, and δ'. The EBS and IBS refer to exon and intron binding sites, respectively. These short sequences are also involved in base-pairing interactions required for intron splicing. The IEP recognizes sequences flanking the central region of the DNA target site. The IEP first recognizes the distal 5' exon region and triggers local DNA unwinding, enabling the formation of base-pairs between the intron and the target site for reverse splicing. Cleavage of the opposite strand occurs after reverse splicing and involves interactions between the IEP and the 3' exon of the target sequence, the most critical being the recognition of T+5.
 Because two crucial segments of the DNA target site are recognized by base paring interactions with the intron RNA, it is possible to change the specificity of the Ll.LtrB intron by modifying the target site recognition sequences in the intron RNA. This flexibility allowed Ll.LtrB to be developed into a highly efficient bacterial gene targeting vector called a TargeTron. Currently, the Targetrons are designed to insert into specific chromosomal loci by a computer algorithm. First, the algorithm uses the probabilistic model to evaluate a DNA sequence for potential intron insertion sites recognized by the IEP and ranks the target sites using a log-odds score. In that process, each target site residue is assigned a value based on the position-specific nucleotide frequency of that residue in a database of active Ll.LtrB group II intron target sites. Next, PCR primers are designed to modify the introns EBS2, EBS1 and δ sequences for insertion into those sites. If the intron is expressed from a donor plasmid, the IBS sequences in the plasmids 5' exon must also complement the introns target site recognition sequences. This is necessary to ensure efficient intron forward splicing and subsequent RNP formation. The required modifications are introduced by a one-step PCR, using three designed primers and one universal primer to mutate the intron at several positions spanning a 350 bp region.
Because two crucial segments of the DNA target site are recognized by base paring interactions with the intron RNA, it is possible to change the specificity of the Ll.LtrB intron by modifying the target site recognition sequences in the intron RNA. This flexibility allowed Ll.LtrB to be developed into a highly efficient bacterial gene targeting vector called a TargeTron. Currently, the Targetrons are designed to insert into specific chromosomal loci by a computer algorithm. First, the algorithm uses the probabilistic model to evaluate a DNA sequence for potential intron insertion sites recognized by the IEP and ranks the target sites using a log-odds score. In that process, each target site residue is assigned a value based on the position-specific nucleotide frequency of that residue in a database of active Ll.LtrB group II intron target sites. Next, PCR primers are designed to modify the introns EBS2, EBS1 and δ sequences for insertion into those sites. If the intron is expressed from a donor plasmid, the IBS sequences in the plasmids 5' exon must also complement the introns target site recognition sequences. This is necessary to ensure efficient intron forward splicing and subsequent RNP formation. The required modifications are introduced by a one-step PCR, using three designed primers and one universal primer to mutate the intron at several positions spanning a 350 bp region.
In addition to site-specific gene inactivation, Targetrons can be used to deliver genes cloned within the introns domain IV to desired chromosomal locations, to repair mutant genes or to generate double stranded breaks and stimulate homologous recombination. In principle, group II intron RNPs can be delivered into mammalian cells to perform targeted genetic repair, although significant technical issues must still be overcome for the development of any new therapeutic approaches.
In bacteria, the Targetron is commonly expressed from a donor plasmid in which the IEP open reading frame (ORF) has been deleted from intron domain IV. The IEP is expressed separately from a position downstream of the 3' exon and intron RNPs are assembled in the host cell. This arrangement enables achieving very high intron mobility frequencies probably due to greater resistance of the ΔORF intron to nucleolytic cleavage in domain IV. The insertion of the ΔORF intron leads to a gene disruption because the intron cannot splice in the absence of the IEP. In principle, the Targetron can be used to obtain either permanent or conditional gene disruption depending on the orientation of the inserted intron with respect to target gene transcription. A Targetron inserted in the sense orientation is transcribed into a functional intron RNA and can be spliced from a precursor RNA to restore the mRNA by providing the IEP in trans, leading to conditional disruption. A Targetron inserted in the antisense orientation is transcribed into non-functional complementary intron sequence and cannot be spliced, yielding a permanent gene disruption.
The major advantage of using Targetrons for gene targeting in bacteria is that computer-designed Targetrons are extremely efficient that allow mutants to be generated without the use of antibiotic resistance markers. This is crucial when the host is naturally resistant to common antibiotics, or for cases in which government regulations restrict the insertion of new antibiotic resistance genes into bioterrorism-related organisms. Also, when multiple knockouts are desired the selection process can be expedited since Targetrons can be introduced sequentially without requiring the intermediate removal of antibiotic resistance markers.
A new Targetron based on the EcI5 group II intron (which was discovered in Escherichia coli virulence plasmid pO157) has recently been developed. The EcI5 recognizes an entirely different target sequence than the LtrB intron, but can again be programmed to insert specifically into desired chromosomal loci. Computer-designed EcI5 Targetrons have been shown to integrate specifically at ten different sites in the E. coli chromosomal lacZ gene at frequencies up to 98% without selection. Based on these and other experiments, the EcI5 Targetron seems to be more efficient that the LtrB Targetron.